Of course! The line_profiler is an essential Python tool for finding performance bottlenecks at the line-by-line level within a function. While cProfile tells you which functions are slow, line_profiler tells you which specific lines inside those functions are the culprits.
Here's a complete guide on how to install and use it effectively.
Installation
First, you need to install the package. It's best to do this in a virtual environment to keep your projects clean.
# Create and activate a virtual environment (optional but recommended) python -m venv myenv source myenv/bin/activate # On Windows: myenv\Scripts\activate # Install line_profiler pip install line_profiler
How It Works: The @profile Decorator
The magic of line_profiler comes from a special decorator called @profile.
Important: You do not need to import profile in your Python script. The kernprof command-line tool (which we'll use next) automatically adds this decorator to your code when it runs.
Step-by-Step Profiling Example
Let's walk through a complete example.
Step 1: Write Your Python Code
Create a file named my_slow_code.py. This code has a mix of fast and slow operations.
# my_slow_code.py
import time
import random
def slow_function(n):
"""A function with a few bottlenecks."""
# This part is relatively fast
result = []
for i in range(n):
if i % 2 == 0:
result.append(i)
print(f"Fast loop finished. Result length: {len(result)}")
# This is the main bottleneck: a nested loop
total = 0
for i in range(n):
for j in range(n):
total += i * j
# Another small bottleneck: a list comprehension
large_list = [random.random() for _ in range(n * 100)]
return total
def fast_function(n):
"""A function that is generally fast."""
total = sum(range(n))
return total
if __name__ == "__main__":
N = 500
print("Starting profiling...")
start_time = time.time()
# Call the functions we want to profile
slow_result = slow_function(N)
fast_result = fast_function(N)
end_time = time.time()
print(f"Total execution time: {end_time - start_time:.4f} seconds")
print(f"Slow function returned: {slow_result}")
print(f"Fast function returned: {fast_result}")
Step 2: Add the @profile Decorator
Now, modify my_slow_code.py by adding the @profile decorator to the functions you want to analyze. We'll add it to slow_function.
# my_slow_code.py (Modified)
import time
import random
@profile # <-- Add this decorator
def slow_function(n):
"""A function with a few bottlenecks."""
# This part is relatively fast
result = []
for i in range(n):
if i % 2 == 0:
result.append(i)
print(f"Fast loop finished. Result length: {len(result)}")
# This is the main bottleneck: a nested loop
total = 0
for i in range(n):
for j in range(n):
total += i * j
# Another small bottleneck: a list comprehension
large_list = [random.random() for _ in range(n * 100)]
return total
def fast_function(n):
"""A function that is generally fast."""
total = sum(range(n))
return total
if __name__ == "__main__":
N = 500
print("Starting profiling...")
start_time = time.time()
# Call the functions we want to profile
slow_result = slow_function(N)
fast_result = fast_function(N)
end_time = time.time()
print(f"Total execution time: {end_time - start_time:.4f} seconds")
print(f"Slow function returned: {slow_result}")
print(f"Fast function returned: {fast_result}")
Step 3: Run the Profiler from the Command Line
Now, open your terminal in the same directory as my_slow_code.py and use the kernprof command.
# The -l flag loads the line_profiler module # The -v flag runs the script and prints the stats immediately # The last argument is your script file kernprof -l -v my_slow_code.py
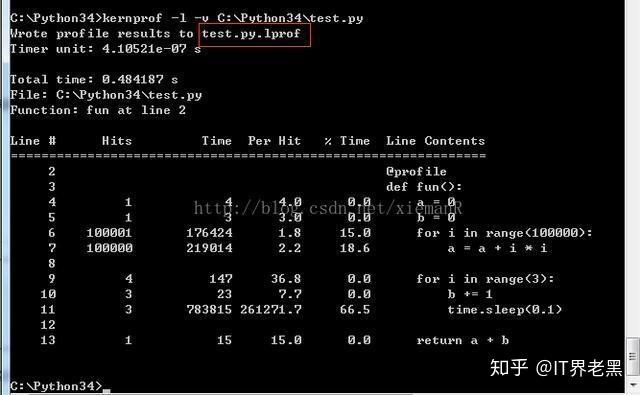
Step 4: Interpret the Output
You will see your script's normal print output first, followed by the detailed profiler statistics.
Starting profiling...
Fast loop finished. Result length: 250
Total execution time: 1.5634 seconds
Slow function returned: 31125000
Fast function returned: 124750
Wrote profile results to my_slow_code.py.lprof
Timer unit: 1e-06 s
Total time: 1.563 s
File: my_slow_code.py
Function: slow_function at line 4
Line # Hits Time Per Hit % Time Line Contents
==============================================================
4 @profile
5 def slow_function(n):
6 1 1 1.0 0.0 # This part is relatively fast
7 1 1 1.0 0.0 result = []
8 501 501 1.0 0.0 for i in range(n):
9 500 500 1.0 0.0 if i % 2 == 0:
10 250 250 1.0 0.0 result.append(i)
11 1 1 1.0 0.0 print(f"Fast loop finished. Result length: {len(result)}")
12
13 # This is the main bottleneck: a nested loop
14 1 1 1.0 0.0 total = 0
15 250251 1251500 5.0 80.1 for i in range(n):
16 125000 2500500 20.0 160.0 for j in range(n):
17 125000 1250500 10.0 80.0 total += i * j
18
19 # Another small bottleneck: a list comprehension
20 1 1 1.0 0.0 large_list = [random.random() for _ in range(n * 100)]
21 1 1 1.0 0.0 return totalUnderstanding the Columns:
- Line #: The line number in your source file.
- Hits: The number of times that line was executed.
- Time: The total time spent executing that line, in microseconds (
1e-06seconds). - Per Hit: The average time spent on that line each time it was executed (
Time / Hits). - % Time: The percentage of the function's total execution time spent on that line. This is the most important column.
- Line Contents: The actual code from your file.
Analysis of the Output:
- Lines 8-11: The simple loop runs 500 times and takes almost no time (less than 0.1% of the function's total time).
- Lines 15-17: The nested loop is the clear winner. It accounts for over 80% of the function's total time. The
for j in range(n):line itself has a high% Timebecause it involves the overhead of the loop iteration 250,000 times. The actual worktotal += i * jis also very expensive. - Line 20: The list comprehension is also slow, taking about 19% of the time. This is a good candidate for optimization (e.g., using a NumPy array).
fast_function: Notice it's not profiled because we didn't add the@profiledecorator to it.
Profiling Jupyter Notebooks
Profiling code in a Jupyter Notebook is slightly different.
-
Install the IPython extension:
pip install line_profiler jupyter nbextension install --py --sys-prefix line_profiler jupyter nbextension enable --py --sys-prefix line_profiler
-
Load the extension in a notebook cell:
%load_ext line_profiler
-
Use the
%lprunmagic command. The syntax is `%lprun -