Of course! Let's dive deep into numpy.linalg.svd(), the primary function for Singular Value Decomposition (SVD) in Python.
What is SVD?
Singular Value Decomposition is a fundamental matrix factorization technique in linear algebra. It breaks down any matrix A (of size m x n) into three other matrices:
A = U Σ Vᵀ
Let's break down each component:
-
U (Left Singular Vectors):
 (图片来源网络,侵删)
(图片来源网络,侵删)- An
m x munitary matrix (orthonormal columns). - Its columns are the eigenvectors of the matrix
A Aᵀ. - They represent the "output" basis vectors or the principal directions in the row space of A.
- An
-
Σ (Sigma - Singular Values):
- An
m x nrectangular diagonal matrix. - The diagonal entries (where
ris the rank of A) are called the singular values. - These values are always non-negative and are typically arranged in descending order (
σ₁ ≥ σ₂ ≥ ... ≥ σᵣ > 0). - They are the square roots of the non-zero eigenvalues of both
A AᵀandAᵀ A. - The singular values represent the "magnitude" or "importance" of each corresponding singular vector.
- An
-
Vᵀ (Right Singular Vectors):
- An
n x nunitary matrix (orthonormal columns).Vᵀis the transpose of V. - Its columns are the eigenvectors of the matrix
Aᵀ A. - They represent the "input" basis vectors or the principal directions in the column space of A.
- An
numpy.linalg.svd()
This function is the workhorse for performing SVD in NumPy.
Syntax
import numpy as np U, s, Vh = np.linalg.svd(a, full_matrices=True, compute_uv=True, hermitian=False)
Parameters
a(array_like): The input matrix, a real or complex array of shape(M, N).full_matrices(bool, optional):- If
True(default),UandVhhave shapes(M, M)and(N, N)respectively. - If
False,UandVhhave shapes(M, K)and(K, N)respectively, whereK = min(M, N). This is more memory-efficient for large, non-square matrices.
- If
compute_uv(bool, optional):- If
True(default), it returnsU,s, andVh. - If
False, it only returns the singular valuess. This can be faster if you only need the singular values.
- If
hermitian(bool, optional):- If
True,ais assumed to be Hermitian (symmetric if real-valued), and the algorithm is optimized for this case. This can be significantly faster.
- If
Return Values
The function returns three arrays: U, s, and Vh.
U: Unitary matrix of shape(M, M)or(M, K).s: The singular values, sorted in descending order. This is a 1-D array, not the full diagonal matrix .Vh: Unitary matrix of shape(N, N)or(K, N). This isVᵀ.
Important Note: The singular values s are returned as a 1-D array. To reconstruct the diagonal matrix , you need to create a zero matrix of the correct shape and place s on its diagonal.
Practical Examples
Example 1: Basic SVD and Reconstruction
Let's decompose a simple matrix and then reconstruct it to verify the decomposition.
import numpy as np
# The original matrix
A = np.array([[1, 2, 3],
[4, 5, 6],
[7, 8, 9],
[10, 11, 12]])
print("Original Matrix A:\n", A)
# Perform SVD
U, s, Vh = np.linalg.svd(A)
print("\nU matrix:\n", U)
print("\nSingular values (s):\n", s)
print("\nVh matrix (V transpose):\n", Vh)
# --- Reconstruction ---
# Create the Sigma matrix from the singular values 's'
# The shape of Sigma should be (m, n)
m, n = A.shape
Sigma = np.zeros((m, n))
# Place the singular values on the diagonal
Sigma[:n, :n] = np.diag(s) # For a non-square matrix, this works well
# Reconstruct A
A_reconstructed = U @ Sigma @ Vh
print("\nReconstructed Matrix A:\n", A_reconstructed)
# Check if the reconstruction is accurate (within floating-point precision)
print("\nIs reconstruction close to original?", np.allclose(A, A_reconstructed))
Example 2: Using full_matrices=False
For a non-square matrix like A (4x3), full_matrices=True gives us a 4x4 U and a 3x3 Vh. We can save memory by using full_matrices=False.
U_small, s_small, Vh_small = np.linalg.svd(A, full_matrices=False)
print("Shape of U with full_matrices=False:", U_small.shape) # Output: (4, 3)
print("Shape of Vh with full_matrices=False:", Vh_small.shape) # Output: (3, 3)
# The reconstruction is still the same
Sigma_small = np.diag(s_small)
A_reconstructed_small = U_small @ Sigma_small @ Vh_small
print("\nReconstruction with small matrices is accurate:", np.allclose(A, A_reconstructed_small))
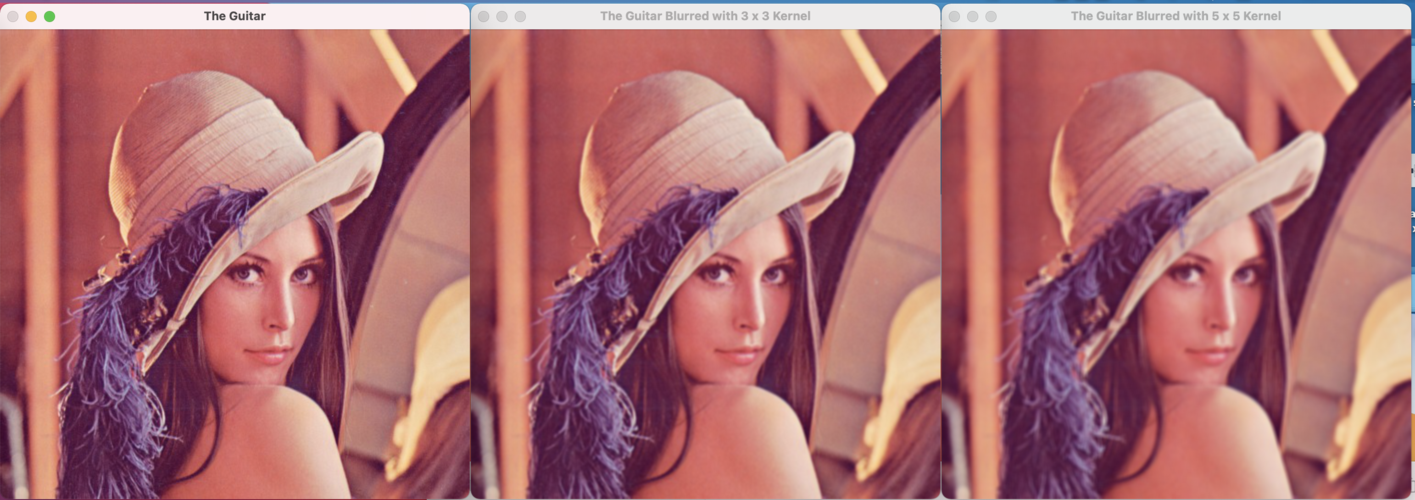
Example 3: Image Compression (A Classic SVD Application)
SVD is powerful for compression because the singular values often decay rapidly. We can keep only the top k singular values and their corresponding vectors to create a low-rank approximation of the original matrix.
import matplotlib.pyplot as plt
from skimage import data
# Load a sample image (a grayscale astronaut image)
image = data.astronaut()
image_gray = np.mean(image, axis=2) # Convert to grayscale
# Perform SVD
U, s, Vh = np.linalg.svd(image_gray)
# Choose the number of components (k) to keep
k = 50
# Create a low-rank approximation
# We take the first k columns of U, first k singular values, and first k rows of Vh
image_compressed = U[:, :k] @ np.diag(s[:k]) @ Vh[:k, :]
# Display the results
fig, axes = plt.subplots(1, 2, figsize=(12, 6))
axes[0].imshow(image_gray, cmap='gray')
axes[0].set_title("Original Image")
axes[0].axis('off')
axes[1].imshow(image_compressed, cmap='gray')
axes[1].set_title(f"Compressed Image (k={k})")
axes[1].axis('off')
plt.show()
# Calculate the compression ratio
original_size = image_gray.shape[0] * image_gray.shape[1]
compressed_size = U[:, :k].shape[0] * U[:, :k].shape[1] + \
k + \
Vh[:k, :].shape[0] * Vh[:k, :].shape[1]
compression_ratio = original_size / compressed_size
print(f"Original size: {original_size} elements")
print(f"Compressed size: {compressed_size} elements")
print(f"Compression ratio: {compression_ratio:.2f}x")
Example 4: Solving a Linear System (Pseudo-Inverse)
For a non-square or singular matrix, the standard inverse doesn't exist. SVD provides a way to compute the Moore-Penrose Pseudo-Inverse, A⁺, which is very useful for solving least-squares problems.
A⁺ = V Σ⁺ Uᵀ
Where is formed by taking the reciprocal of non-zero singular values and then transposing the result.
# An overdetermined system (more equations than unknowns)
# Ax = b, where A is 4x2 and b is 4x1
A_sys = np.array([[1, 2],
[3, 4],
[5, 6],
[7, 8]])
b_sys = np.array([10, 20, 30, 40])
# Solve using SVD and the pseudo-inverse
U_sys, s_sys, Vh_sys = np.linalg.svd(A_sys, full_matrices=False)
# Create the pseudo-inverse of Sigma
Sigma_pseudo = np.diag(1 / s_sys)
# Calculate the pseudo-inverse of A
A_pseudo_inv = Vh_sys.T @ Sigma_pseudo @ U_sys.T
# Solve for x
x_solution = A_pseudo_inv @ b_sys
print("Matrix A:\n", A_sys)
print("\nVector b:\n", b_sys)
print("\nSolution x (using SVD pseudo-inverse):\n", x_solution)
# Compare with numpy's built-in lstsq (least-squares solution)
x_lstsq, _, _, _ = np.linalg.lstsq(A_sys, b_sys, rcond=None)
print("\nSolution x (using np.linalg.lstsq):\n", x_lstsq)
Common Pitfalls and Key Takeaways
VhisVᵀ, notV: The function returnsV transpose. If you need the matrix V itself, you must transpose the result:V = Vh.T.sis 1-D: The singular values are returned as a flat array. You must manually construct the diagonal matrix for reconstruction.- Floating-Point Precision: Due to floating-point arithmetic, the reconstructed matrix might not be exactly the same as the original. Always use
np.allclose(A, A_reconstructed)for comparison, notA == A_reconstructed. - Rank and Zero Singular Values: The number of non-zero singular values in
sgives you the rank of the matrixA. This is a numerically stable way to find rank. full_matrices: For large datasets, always considerfull_matrices=Falseto save memory and computation time, especially if you only need the first few singular vectors.